New method to determine the risk of skin cancer relapse
BMBF-funding for MelEVIR
How likely do patients develop cancer again after a tumor has been surgically removed? This is the central question of the new research project „MelEVIR – Melanoma, Extracellular Vesicles, and Immune Response“ that will be funded by the German Ministry of Education and Research (BMBF) with overall 1.3 million Euro. The project started in April 2016 with a duration of three years.
To answer this question, researchers at the Department of Dermatology (Prof. Dr. Vera González (Coordinator), Prof. Dr. A. Baur, Prof. Dr. L. Heinzerling), at the Chair of Medical Informatics (Prof. Dr. H.U. Prokosch, Dr. T. Ganslandt) as well as the joint researcher of the University of Rostock, Prof. Dr. O. Wolkenhauer (Department of Systems Biology and Bioinformatics) have a closer look at the so-called “minimal residual disease” (MRD) after primary surgery in highly metastatic tumors like melanoma.
Even after successful tumor removal, a relapse can occur many years later. The individual risk for a relapse varies a lot. Therefore, physicians focus more and more on detecting very early tumor growth or MRD. The MRD consists of residual tumor cells and micro-metastases, which could be dormant for years and even decades before they eventually re-emerge as solid metastases. At present, the early detection of tumor growth is typically monitored by the assessment of tumor markers (not always sufficiently specific and sensitive) or using imaging systems like the PET (expensive and labor-intensive). In everyday clinical practice, these approaches contribute fairly little to detecting very early tumor growth or MRD. Thus, there is a high and urgent demand for simpler and more sensitive diagnostic procedures.
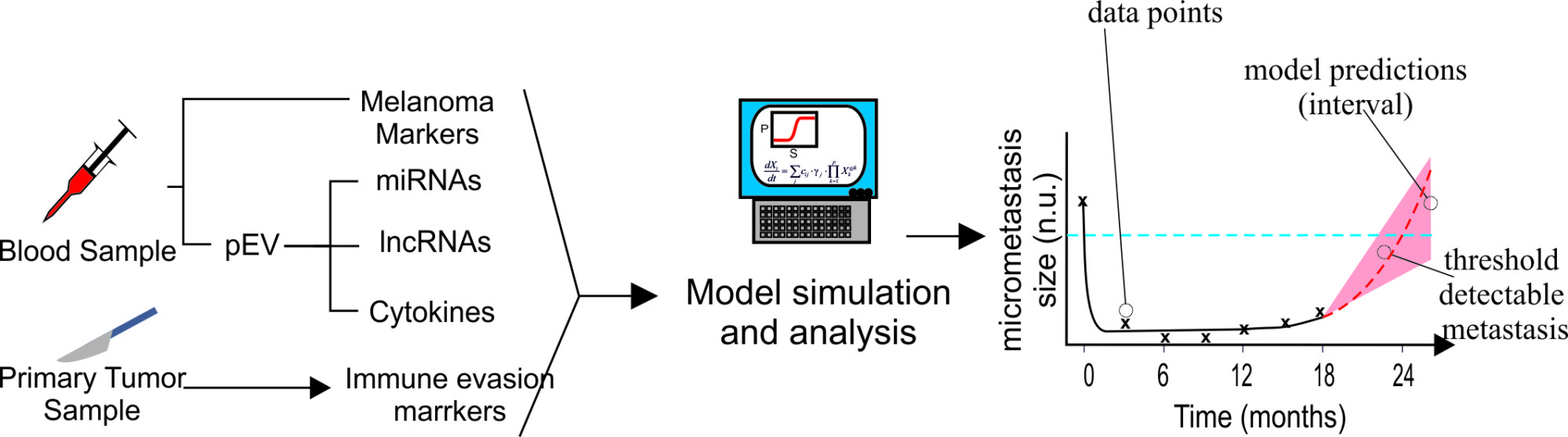
A systems methodology to assess the risk of tumor relapse in melanoma by the profiling of plasma extracellular vesicles
Extracellular vesicles (EV) are cell-derived vesicles which are shed into plasma (pEV), urine and other biological fluids, as well as into cell culture supernatants. Many cell types including immune cells and tumor cells secrete Es carrying a high content of functionally active proteins and different types of RNA, including miRNAS. Recent publications show significantly elevated levels of extracellular vesicles in serum of cancer patients, and many suggest this could be used for accurate diagnostics of tumor relapse. In preliminary work, the group has found that tumor-bearing melanoma patients have significantly elevated levels of pEV as compared to control groups. Importantly, 50% of operated high-risk patients showed similarly elevated profiles as tumor-bearing patients.
The aim of the project is to develop, test and prepare for translation into clinical practice a systems-biology-based diagnostic tool for assessing the probability of tumor relapse in melanoma patients, based on the profiling of pE. Systems Biology is a sophisticated methodological approach that combines quantitative experimental data, mathematical modeling and simulation and tools from computational biology to address biological and biomedical questions.
In the methodology proposed in the project, in vitro and clinical data are integrated using data-driven mathematical modeling. The insights obtained from patient data analysis, reconstruction of biochemical networks, and model simulations are used to a) select a set of microRNA, long non-coding RN, and proteins present in pEs of patients to be measured in a blood test as surrogates of immune system activity against MRD and b) assess the probability of tumor relapse in the close future.
This project proposal puts together efforts from modelers, bioinformaticians, and biomedical researchers to develop, test and prepare for translation into clinical practice a data-driven, mathematical-model-based diagnostic tool for assessment of the probability of tumor relapse in melanoma patients. The expected results will allow individualized diagnostics and treatment in melanoma patients. Here, the three branches of the computational systems biology (high-throughput data analysis, bioinformatics and mathematical modeling) are merged into a systems biology workflow for the iterative integration of in vitro and clinical data. Projects like this try to pave the way towards a systems-based translational and personalized medicine, in line with the economic and scientific strategy outlined by German and European scientific committees to promote a biotechnology- and knowledge-funded economy.
Further information
Prof. Dr. Julio Vera González
Phone: 09131/85-45876